Stopping a Runaway Train – How Bacteria Avoid Making Unwanted RNA
Publication in Science by biochemists from Freie Universität Berlin and international colleagues
№ 235/2020 from Dec 03, 2020
An important gene expression process in bacteria seems to proceed differently than described in textbooks. This is the result of an international team of scientists headed by the Structural Biochemistry group at Freie Universität Berlin. Their work deals with, among other things, the question of why pathogenic bacteria turn out to be harmful to us without harming themselves. The findings were published in the journal Science (https://doi.org/10.1126/science.abd1673).
All life on Earth uses RNA molecules as copies of the genetic instructions in DNA to produce proteins that mediate cellular processes. These RNA molecules are synthesized by RNA polymerases, which are complex and powerful molecular machines, in a process called transcription. Transcription is an assembly line-like process: During each duty cycle, the RNA polymerase picks up a building block of the DNA chain, finds the corresponding RNA building block, and attaches it to the growing RNA chain. “The polymerase can tirelessly form RNA chains of up to a million building blocks. However, it cannot tell whether the RNA molecule it is making is crucial, useless, or even harmful for the cell,” says Professor Markus Wahl, one of the corresponding authors of the study and an expert in structural biology at Freie Universität.
When RNA polymerases transcribe useless or harmful sequences of DNA, rather than focus on the DNA sequences that are most important for the cell at a given time, they often do not encounter stop signals in the DNA that normally terminate the legitimate transcription processes. Therefore, such “off-road” RNA polymerases need to be switched off by proteins called termination factors. These termination factors cause the RNA polymerase to detach from its DNA template and its RNA product; the so-called transcription complex is disrupted.
Bacterial Rho protein was the first transcription termination factor identified. Rho is a “motor protein” that can walk on RNA and that exerts mechanical forces that are much stronger than those exerted by RNA polymerase. These and other observations led to the prevailing hypothesis that the Rho protein binds certain recognition sequences on the RNA, then uses its powerful motor to catch up to RNA polymerase and pulls the RNA chain out of the transcription complex. “However, this model could never explain how Rho can terminate the synthesis of RNAs that lack such entry sequences,” says Professor Irina Artsimovitch, a biochemist at The Ohio State University in Columbus (USA) and another senior researcher on the study. “These include RNA transcripts of virulence genes, the expression of which is too costly for bacteria, except during a successful infection of a host organism.” The ability of the Rho protein to switch off transcription is therefore essential for bacteria to successfully survive in dormant states until there is an opportunity to attack a host. However, to achieve such precise timing, Rho has to stop transcription very quickly. So far, this has made it difficult to experimentally observe Rho at work.
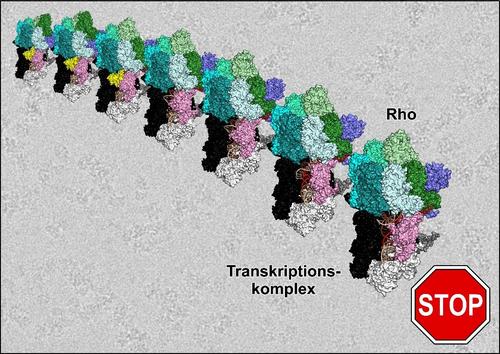
A team of scientists from Berlin, together with colleagues from The Ohio State University, the Center for DNA Fingerprinting and Diagnostics in Hyderabad, India, and the University of Turku, Finland, have now made observations that call the above textbook model into question. Dr. Nelly Said and Dr. Tarek Hilal from the Structural Biochemistry group at Freie Universität Berlin succeeded in producing a transcription complex in the process of termination mediated by the Rho protein, and in elucidating molecular 3D structures of the complex at high resolution, using cryogenic single-particle electron microscopy (cryo-EM). They determined the structures of seven states that appear to represent sequential steps in termination: from Rho recruitment to inactivation of the RNA polymerase (see figure). “Cryo-EM is uniquely capable of capturing an ensemble of even unstable, intermediate biomolecular structures. It is therefore an ideal method for investigating dynamic processes, such as the fast but still highly regulated termination of transcription,” explain Said and Hilal.
According to the biochemists, the structures show that the Rho protein does not work as it has long been assumed. Instead of initially binding to RNA, Rho first docks to the polymerase and two other proteins, called NusA and NusG. Instead of pulling on RNA, Rho pushes on RNA polymerase and induces many rearrangements in the enzyme. The polymerase is thus forced to loosen its grip on the DNA. The Rho protein also causes the RNA to move out of the active site of the RNA polymerase, effectively stopping RNA elongation and thus turning off the transcription machinery. “Most remarkable, however, is that Rho elicits these effects without using its strong motor activity,” says Professor Wahl.
While cryo-EM can reveal the various conformations of a biomolecular complex, it cannot directly explain the precise functional meanings of these states. Therefore, the scientists used complementary biochemical and genetic experiments to fully validate the predictions of the alternative model. The results show that Rho-mediated termination is a team effort, in which the Nus proteins ensure that Rho is only called into action when needed, and that no matter how fast the RNA polymerase works, it cannot escape Rho that rides along on the transcription complex. “In hindsight, these results are not at all surprising,” explains Professor Artsimovitch. “By hitchhiking on RNA polymerase from the start instead of trying to catch up later, Rho can ensure that only the desired RNAs are produced.”
“Due to the robotic nature of RNA polymerases, unwanted transcription is a problem for all forms of life, including humans. It is therefore likely that similar mechanisms for quality control of transcription exist in all living beings,” says Wahl.
Figure
Cryo-EM structures showing the Rho protein on path to terminating a transcription complex.
Image Credit: Markus Wahl, Freie Universität Berlin
The image may be downloaded by journalists. Use of the image within the context of this press release and with proper citation is free of charge.
Further Information
Publication
Said N, Hilal T, Sunday ND, Khatri A, Bürger J, Mielke T, Belogurov GA, Loll B, Sen R, Artsimovitch I, Wahl MC (2020) Steps toward translocation-independent RNA polymerase inactivation by terminator ATPase ρ. https://doi.org/10.1126/science.abd1673
Contact
Prof. Irina Artsimovitch, PhD, The Ohio State University, Department of Microbiology and Center for RNA Biology, Columbus, OH, USA; https://microbiology.osu.edu/people/artsimovitch.1; Email: artsimovitch.1@osu.edu
Prof. Markus C. Wahl, PhD, Freie Universität Berlin, Institut für Chemie und Biochemie, Laboratory of Structural Biochemistry, https://www.bcp.fu-berlin.de/en/chemie/biochemie/research-groups/wahl-group, Email: markus.wahl@fu-berlin.de